
ncert biology class 12 pdf free download, ncert biology book class 12 pdf free download, ncert biology class 11 pdf free download, ncert books pdf free download, ncert biology class 12 pdf, ncert books free download pdf, ncert books download pdf free, biology ncert class 12 pdf, download ncert books pdf, ncert books download pdf, ncert maths book class 12 pdf, ncert book download pdf, ncert pdf books, ncert books download pdf format , biotechnology principles and processes 12 notes, class 12 biology notes, biotechnology principles and processes class 12, biotechnology principles and processes class 12 notes, class 12 biotechnology principles and processes, note biology, biology notes, biotechnology principles and processes, class 12 cbiology chapter 11 notes, 12th standard biology
is a broad area of science involving multiple disciplines designed to use living organisms or their products to perform valuable industrial or manufacturing processes or applications pertaining to human benefit.
An organism’s genome contains virtually all the information necessary for its growth and development
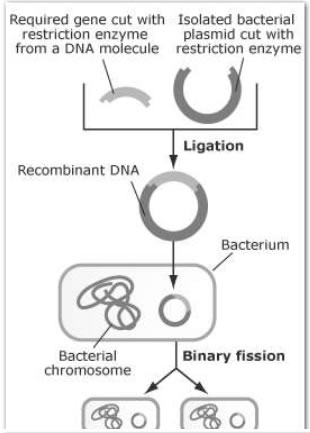
1. The required gene is cut from a DNA molecule using a restriction enzyme.
2. A bacterial plasmid is isolated and cut with the same restriction enzyme. This ensures cut ends are complementary (same base sequence) to the ends of the required gene.
3. The required gene is joined to the plasmid using the enzyme DNA ligase in a process called ligation.
4. The resulting recombinant plasmid is returned to the bacterial cell.
5. The bacteria reproduce and the required gene is cloned
.
Quite straight forward to isolate DNA
For instance, to isolate genomic DNA
1. Remove tissue from organism
2. Homogenize in lysis buffer containing guanidine thiocyanate (denatures proteins)
3. Mix with phenol/chloroform - removes proteins
4. Keep aqueous phase (contains DNA)
5. Add alcohol (ethanol or isopropanol) to precipitate DNA from solution
6. Collect DNA pellet by centrifugation
7. Dry DNA pellet and resuspend in buffer
8. Store at 4°C
Each cell (with a few exceptions) carries a copy of the DNA sequences which make up the organism’s
genome.
How do we manipulate DNA?
It used to be difficult to isolate enough of a particular DNA sequence to carry out further manipulation
and/or characterization of its molecular sequence
| Techniques for | |
| - Isolation | |
| - Digestion | |
| - Fractionation | |
| - Purification of the TARGET fragment | |
| - Cloning into vectors | |
| - Transformation of host cell and selection | |
| - Replication | |
| - Analysis | |
Introduction of recombinant DNA into host cells:
Some commonly used procedures:
1. Transformation
2. Transfection
3. Electroporation
4. Biolistics
5. Agrobacterium mediated gene transfe
DNA is manipulated using various enzymes that modify and/or synthesise it Until 1970 there were no convenient methods available for cutting DNA into discrete, manageable fragments.
1970 - The Beginning of the Revolution Discovery of a restriction enzyme in the bacterium Haemophilus influenzae
• Restriction enzymes are endonucleases
•Bacterial enzymes.
•Different bacterial strains express different restriction enzymes.
•The names of restriction enzymes are derived from the name of the bacterial strain
they are isolated from.
•Cut (hydrolyse) DNA into defined and REPRODUCIBLE fragments.
•Basic tools of gene cloning .
Titles of restriction enzymes are derived from the first letter of the genus + the first two letters of the species of organism from which they were isolated.
This is known as a Restriction Site The phosphodiester bond is cleaved between specific bases, one on each DNA strand
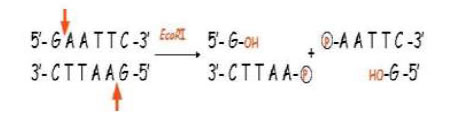
The product of each reaction is two double stranded DNA fragments
Restriction enzymes do not discriminate between DNA from different organisms
Restriction endonucleases are a natural part of the bacterial defence system
• Part of the restriction/modification system found in many bacteria
• These enzymes RESTRICT the ability of foreign DNA (such as bacteriophage DNA) to
infect/invade the host bacterial cell by cutting it up (degrading it)
• The host DNA is MODIFIED by METHYLATION of the sequences these enzymes
recognise
o Methyl groups are added to C or A nucleotides in order to protect the bacterial
host DNA from degradation by its own enzymes
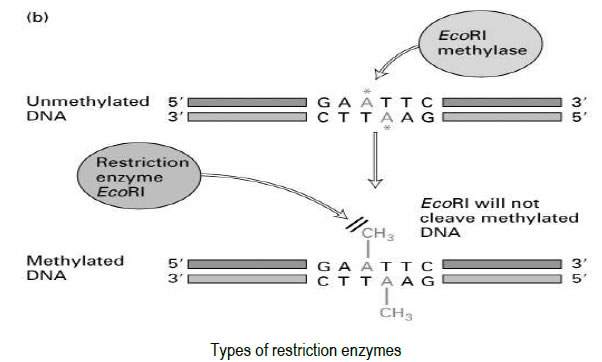
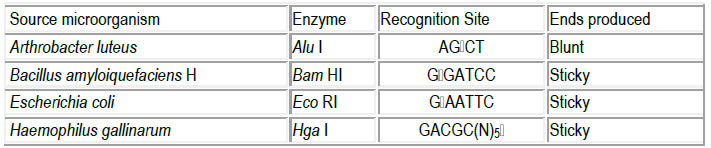
• Type I Recognise specific sequences but then track along DNA (~1000-5000 bases)
before cutting one of the strands and releasing a number of nucleotides (~75) where
the cut is made. A second molecule of the endonuclease is required to cut the 2nd
strand of the DNA
o e.g. EcoK.
o Require Mg2+, ATP and SAM (S-adenosyl methionine) cofactors for function
Type II Recognise a specific target sequence in DNA, and then break the DNA (both strands), within or
close to, the recognition site.Only they are used in rDNA technology as they recognize and cut DNA
within a specific sequence typically consisting of 4-8 bp.
o e.g. EcoRI
o Usually require Mg2+
• Type III Intermediate properties between type I and type II. Break both DNA strands at
a defined distance from a recognition site
o e.g. HgaI
o Require Mg2+ and ATP
Hundreds of restriction enzymes have been isolated and characterised
• Enables DNA to be cut into discrete, manageable fragments
• Type II enzymes are those used in the vast majority of molecular biology techniques
• Many are now commercially available
Many Type II restriction endonucleases recognise PALINDROMIC sequences (From Greek
palindromos, running back again, recurring: palin, again)
A segment of double-stranded DNA in which the nucleotide sequence of one strand reads in reverse
order to that of the complementary strand. (Always read from the same direction)
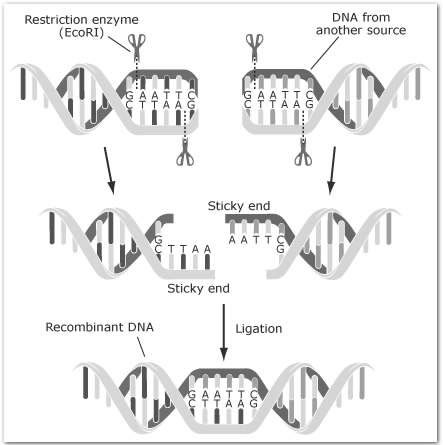
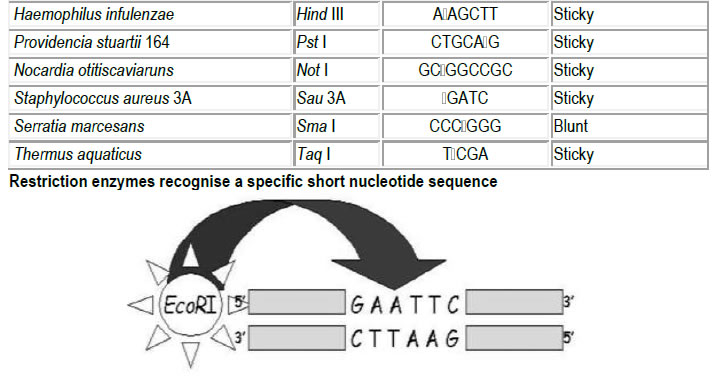
For example, EcoRI recognises the sequence
5’-G A A T T C-3’
3’-C T T A A G-5’
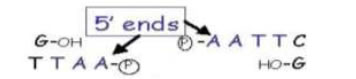
Different enzymes cut at different positions and can create single stranded ends (’sticky ends’)
• Some generate 5’ overhangs - eg: EcoRI
• Some generate 3’ overhangs - eg: PstI
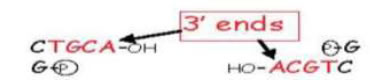
• Some generate 3’ overhangs - eg: PstI

Examples of restriction enzymes and the sequences they cleave
• The single stranded termini (or ends) can base pair (ANNEAL) with any complementary single stranded termini
This is the basis for RECOMBINANT DNA TECHNOLOGY
• Inserting foreign DNA into a cloning vector
After ligating a particular DNA sequence into a cloning vector, it is necessary to check that the correct
fragment has been taken up. Sometimes it is also necessary to ensure that the foreign DNA sequence
is in a certain orientation relative to sequences present in the cloning vector.
• Checking the size of the insert
• Checking the orientation of the insert
• Determining pattern of restriction sites within insert DNA
Separation of DNA fragments in order to isolate and analyse DNA cut by restriction enzymes
Electrophoresis is a technique used to separate and sometimes purify macromolecules - especially
proteins and nucleic acids - that differ in size, charge or conformation. When charged molecules are
placed in an electric field, they migrate toward either the positive or negative pole according to their
charge.
DNA is electrophoresed through the agarose gel from the cathode (negative) to the anode(positive)
when a voltage is applied, due to the net negative charge carried on DNA.
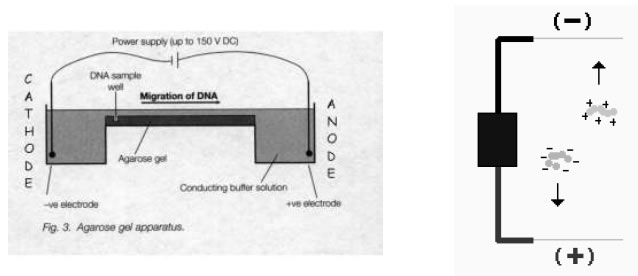
When the DNA has been electrophoresed, the gel is stained in a solution containing the chemical ethidium bromide. This compound binds tightly to DNA and fluoresces strongly under UV light - allowing the visualisation and detection of the DNA.
Recombinant DNA: Plasmids, cloning
What is DNA cloning?
DNA cloning is the isolation of a fragment or fragments of DNA from an organism and placing in a VECTOR that replicates independently of chromosomal DNA. The RECOMBINANT DNA is propagated in a host organism; the resulting CLONES are a set of genetically identical organisms which contain the recombinant DNA
1) DNA sequencing
2) Protein production
3) Engineering animals/plants/proteins
Isolated DNA is cloned into VECTORS for long term storage, propagation of the DNA and for production of protein from gene(s) encoded in the DNA
Cloning vectors are extra-chromosomal ’replicons’ of DNA which can be isolated and can replicate independently of the chromosome. Vectors usually contain a selectable marker - a gene that allows selection of cells carrying the vector e.g. by conferring resistance to a toxin. DNA of interest can be cloned into the vector and replicated in host cells, usually one which has been well characterised.
• Bacterial plasmids
• Bacteriophages
• Cosmids
• Yeast artificial chromosomes (YACs)
• Ti plasmid (plants)
• Eukaryotic viruses such as baculovirus (insect cells), SV40 virus and retroviruses.
This process marks autonomous replication in vector. ORI is a specific sequence of nucleotide in DNA from where replication starts. When foreign DNA is linked to this sequence then along with vector replication, foreign (desirable) DNA also starts replicating within host cell.
Charecteristics of Selectable marker: A gene whose expression allows one to identify cells that have been transforrned or transfected with a vector containing the marker gene. A marker gene is used to determine if a piece of DNA has been successfully inserted into the host organism. Gene usually encoding resistance to an antibiotic. A selectable marker will protect the organism from a selective agent that would normally kill it or prevent its growth.
Restriction sites
Allow cleavage of specific sequence by specific Restriction Endonuclease. Restriction sites in
E.coli cloning vector pBR322 include HindIII , EcoRI , BamHI , SalI, PvuI, PstI, ClaI etc.
Refer NCERT text book diagram of pBR322
A Cloning Vector that Works with Plant Cells
Most commonly used plant cloning vector "Ti" plasmid, or tumor-inducing plasmid. Found in cells of
the bacterium known as Agrobacterium tumefaciens, normally lives in soil. Bacterium has ability to
infect plants and cause a crown gall, or tumorous lump, to form at the site of infection.
Ti plasmid - called T DNA - separates from the plasmid and incorporates into the host cell genome. This
aspect of Ti plasmid function has made it useful as a plant cloning vector (natural genetic engineer).
Plasmids are the most commonly used vector system. Several types available for cloning of foreign
DNA in the host organism Escherichia coli. Many E. coli plasmids allow the expression of proteins
encoded by the cloned DNA
Bacteriophage another common vector system used for cloning DNA. These are viruses which ’infect’
E. coli. The M13 bacteriophage is a single-stranded DNA virus which replicates in E. coli in a doublestranded
form that can be manipulated like a plasmid. It can be used to produce single-stranded DNA
copies which are useful for DNA sequencing.
Bacteriophage common vector system used to make DNA libraries. It allows the cloning of larger
fragments of DNA than can be incorporated into plasmids.
Transformation is the process by which plasmids (or other DNA) can be introduced into a cell. For E. coli transformation with plasmids is quite straightforward. Plasmids can be introduced by electroporation or by incubation in the presence of divalent cations (usually Ca2+) and a brief heat shock (42°C) which induces the E. coli cells to take up the foreign DNA
1. Two antibiotic selection and replica plating
2. Color selection: blue/white selection using the lacz gene
Subcloning a DNA fragment into an active gene (usually a marker gene whose function can be easily detected) will disrupt the function of that gene. This can be detected by looking for colonies that no longer display that phenotype.
A more common method to determine which transformants contain plasmids with inserts is to use colour selection. For E. coli, this involves the lac complex and blue/white screening.
Colonies carrying plasmid with no insert will be coloured blue whereas colonies carrying recombinant
plasmid will be white
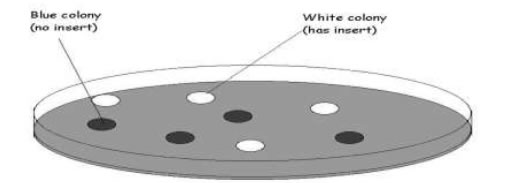
For plasmids such as pBR322, which contains two antibiotic resistance genes, cloning an insert into one of these will disrupt that gene and inactivate the resistance to that antibiotic.
Analysing complex nucleic acid mixtures (DNA or RNA)
The total cellular DNA of an organism (genome) or the cellular content of RNA are complex mixtures of
different nucleic acid sequences. Restriction digest of a complex genome can generate millions of
specific restriction fragments and there can be several fragments of exactly the same size which will not
be separated from each other by electrophoresis.
Techniques have been devised to identify specific nucleic acids in these complex mixtures
• Southern blotting - DNA
• Northern blotting - RNA
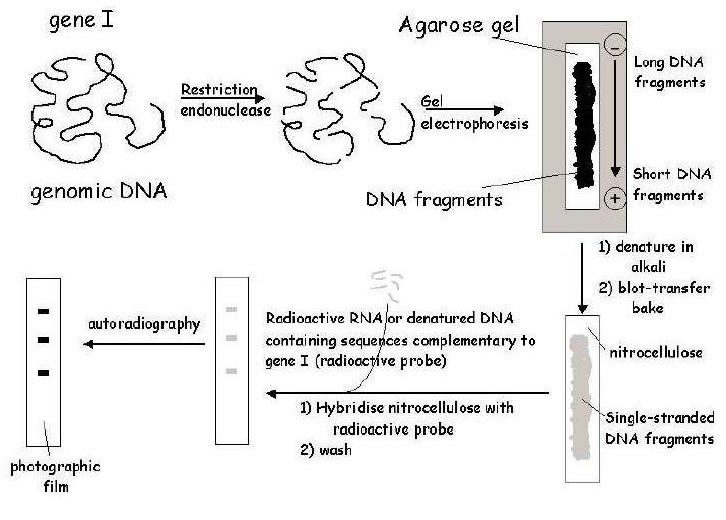
Technique devised by Ed Southern in 1975, is a commonly used method for the identification of DNA fragments that are complementary to a know DNA sequence. Allows a comparison between the genome of a particular organism and that of an available gene or gene fragment ( probe). It can tell us whether an organism contains a particular gene (DNA fragment) or not
1. Chromosomal DNA is isolated from the organism of interest, and digested to completion with a
restriction endonuclease enzyme.
2. The restriction fragments are then subjected to electrophoresis on an agarose gel, which separates
the fragments on the basis of size.
3. DNA fragments in the gel are denatured (i.e. separated into single strands) using an alkaline solution.
4 .Transfer fragments from the gel onto nitrocellulose filter or nylon membrane.
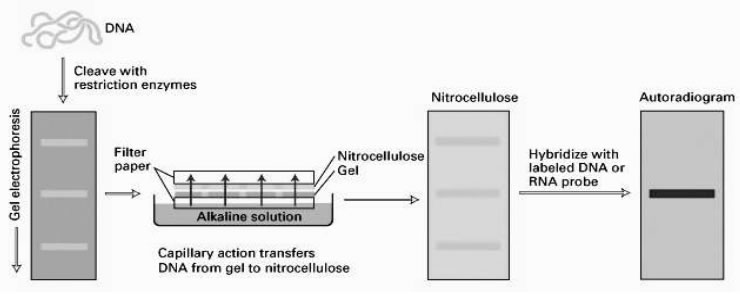 Fig 7-32, Lodish et al (4th ed.)
Fig 7-32, Lodish et al (4th ed.)
DNA is bound irreversibly to the filter/membrane by baking at high temperature (nitrocellulose) or crosslinking through exposure to UV light (nylon).
Final step is to immerse the membrane in a solution containing the probe - either a DNA (cDNA clone, genomic fragment, oligonucleotide or RNA ) can be used. This is DNA hybridisation The membrane is washed to remove non-specifically bound probe, and is then exposed to X-ray film - a process called autoradiography. The principle of Southern blotting
PCR is a technique for the in vitro amplification of a desired sequence of DNA. PCR allows the
generation of a large quantity of DNA product (up to several
• g) from only a few starting
copies. it has been shown that PCR can be used to generate a detectable quantity of DNA from only one starting
target (or template) molecule.
PCR developed in the mid-1980, has found multiple applications, such as :-
1. Rapid amplification of intact genes or gene fragments
2. Generation of large amounts of DNA for sequencing
3. Generation of probes specific for uncloned genes by selective amplification of a
specific segment of cDNA
4. Analysis of mutations for medical applications
5. Detection of minute amounts of DNA for forensic purposes
6. Amplification of chromosomal regions adjacent to genes of known sequence and many
more·
Development of PCR won the Nobel prize for Kary Mullis and co-workers.
PCR reaction is a DNA synthesis reaction that depends on the extension of primers annealed to opposite strands of a dsDNA template that has been denatured (melted apart) at temperatures near boiling. By repeating the melting, annealing and extension steps, several copies of the original template DNA can be generated.
The amount of starting material (target) needed is very small
Not necessary to isolate the desired sequence, because it will be defined by the primers that are used in the reaction. The primers are oligonucleotides complementary to different regions on the 2 strands of DNA template (flanking the region to be amplified). The primer acts as a starting point for DNA synthesis. The oligo is extended from its 3’ end by DNA

PCR is a cycle of three steps:
1. DENATURATION - the strands of the DNA are melted apart by heating to 95°C
2. ANNEALING - the temperature is reduced to ~ 55°C to allow the primers to anneal to
the target DNA
3. POLYMERISATION / EXTENSION - the temperature is changed to the optimum temperature for the DNA polymerase to catalyse extension of the primers, i.e. to copy
the DNA between the primers.
The cycle is repeated over and over again - as many times as needed to produce a detectable amount of product.
The breakthrough came with the discovery of the thermostable DNA polymerase Taq
polymerase, from the thermophilic bacterium, Thermus aquaticus, which lives in hot springs.
Taq polymerase enzyme can resist high temperatures required to melt the template DNA apart
without denaturation (loss of activity) and works best at high temperatures (72°C). This led to
improved specificity & sensitivity. Annealing of primers to sites other than the target sequence is
significantly reduced at the higher temperatures used for Taq polymerase.
Applications of PCR
1) Cloning a gene encoding a known protein
2) Amplifying ’old DNA’
3) Amplifying cloned DNA from vectors
4) Creating mutations in cloned genes
5) Rapid amplification of cDNA ends - RACE
6) Detecting bacterial or viral infection
* AIDs infection
* Tuberculosis (Mycobacterium tuberculosis)
7) Cancer Detecting mutations that occur in cancer and monitoring cancer therapy. Determining if a patient is free
of malignant cells
8) Genetic diagnosis
* Cystic fibrosis
* Muscular dystrophy
* Haemophilia A and B
* Sickle cell anemia
certain cancers are caused by specific and reproducible mutations: eg. Retinoblastoma - childhood cancer of the eye. The heritable form (germ line mutation of one of the two retinoblastoma allelles): mutation is detected in all cells. Spontaneous form: only detected in tumour tissue.
eg determining the sex of foetus for those at risk of X-linked disorders
PCR is one of the most versatile techniques invented, and has so many applications that this list could
go on for quite some time.
It refers to the recovery and purification of biosynthetic products, particularly pharmaceuticals, from natural sources such as animal or plant tissue or fermentation broth Stages in Downstream Processing A widely recognized heuristic for categorizing downstream processing operations divides them into four groups which are applied in order to bring a product from its natural state as a component of a tissue, cell or fermentation broth through progressive improvements in purity and concentration.
Removal of insoluble Product Isolation Product Purification Product Polishing
Adult stem cells
The stem cells found in a developed organism and have the twin properties of self-renewal and
differentiation. These can be obtained from fetal cord blood and bone marrow. They are multipotent in nature.
An increase in the number of copies of a specific DNA fragment; can be in vivo or in vitro. See also :cloning, polymerase chain reaction
Adding pertinent information such as gene coded for, amino acid sequence, or other complementary to the database entry of raw sequence of DNA bases.
Nucleic acid that has a sequence exactly opposite to an mRNA molecule made by the body; binds to the mRNA molecule to prevent a protein from being made.
RNA technology An RNA molecule that is the reverse complement of a naturally occurring mRNA, and which can be used to prevent translation of that mRNA in a transformed cell.
A technique that uses X-ray film to visualize radioactively labeled molecules or fragments of molecules; used in analyzing length and number of DNA fragments after they are separated by gel electrophoresis.
A vector used to clone DNA fragments (100 to 300 kb insert size; average, 150 kb) in Escherichia coli cells. Based on naturally occurring F-factor plasmid found in the bacterium E. coli.
The order of nucleotide bases in a DNA molecule; determines structure of proteins encoded by that DNA.
The science of managing and analyzing biological data using advanced computing techniques. Especially important in analyzing genomic research data.
Remarkable method developed to introduce foreign DNA into mainly plant cells is by using a gene or particle gun. Microscopic particles of gold or tungsten are coated with the DNA of interest and bombarded onto cells with a device much like a particle gun. Hence the term biolistics is used.
Set of biological techniques developed through basic research and now applied to research and product development. In particular, biotechnology refers to the use by industry of recombinant DNA, cell fusion, and new bioprocessing techniques.
Diseases in which abnormal cells divide and grow unchecked. Cancer can spread from its original site to other parts of the body and can be fatal. See also:hereditary cancer, sporadic cancer
Something which causes cancer to occur by causing changes in a cell’s DNA. See also:mutagen
An individual who possesses an unexpressed recessive trait.
library A collection of DNA sequences that code for genes. The sequences are generated in the laboratory from mRNA sequences. See also: messenger RNA
An exact copy made of biological material such as a DNA segment (eg. a gene or other region), a whole cell, or complete organism.
Using specialized DNA technology to produce multiple, exact copies of a single gene or other segment of DNA to obtain enough material for further study. Process, used by researchers in the Human Genome Project, referred to as cloning DNA. Resulting cloned (copied) collections of DNA molecules constitute clone libraries. Second type of cloning exploits the natural process of cell division to make many copies of an entire cell. The genetic makeup of these cloned cells, called cell line, is identical to the original cell. Third type of cloning produces complete, genetically identical animals such as the famous Scottish sheep, Dolly.
DNA molecule originating from a virus, a plasmid, or the cell of a higher organism into which another DNA fragment of appropriate size can be integrated without loss of the vector’s capacity for selfreplication; vectors introduce foreign DNA into host cells, where the DNA can be reproduced in large quantities. Examples are plasmids, cosmids, and yeast artificial chromosomes; vectors are often recombinant molecules containing DNA sequences from several sources.
DNA that is synthesized in the laboratory from a messenger RNA template.
Nucleic acid base sequence that can form a double-stranded structure with another DNA fragment by following base-pairing rules (A pairs with T and C with G). The complementary sequence to GTAC for example, is CATG.
Artificially constructed cloning vector containing the cos gene of phage lambda. Cosmids can be packaged in lambda phage particles for infection into E. coli; Permits cloning of larger DNA fragments (up to 45kb) than can be introduced into bacterial hosts in plasmid vectors. DNA bank A service that stores DNA extracted from blood samples or other human tissue.
A PCR technique that determines the alleles present at different STR (short tandem repeat) loci within a genome in order to use DNA information to identify individuals.
Genes encoding proteins that correct errors in DNA sequencing.
The use of existing DNA as a template for the synthesis of new DNA strands. In humans and other eukaryotes, replication occurs in the cell nucleus.
The relative order of base pairs, whether in a DNA fragment, gene, chromosome, or an entire genome. See also: base sequence analysis
The twisted-ladder shape that two linear strands of DNA assume when complementary nucleotides on opposing strands bond together.
A method of separating large molecules (such as DNA fragments or proteins) from a mixture of similar molecules. An electric current is passed through a medium containing the mixture, and each kind of molecule travels through the medium at a different rate, depending on its electrical charge and size. Agarose and acryl amide gels are the media commonly used for electrophoresis of proteins and nucleic acids.
A process using high-voltage current to make cell membranes permeable to allow the introduction of new DNA; commonly used in recombinant DNA technology. See also:transfection Embryonic stem (ES) cells An embryonic cell having totipotency that can replicate indefinitely, transform into other types of cells, and serve as a continuous source of new cells. These cells are derived from inner cell mass of the blastocyst or the 4-8 cell stage of embryo.
See:restriction enzyme
Common bacterium that has been studied intensively by geneticists because of its small genome size, normal lack of pathogenicity, and ease of growth in the laboratory.
DNA originating outside an organism that has been introduced into the organism.
The protein-coding DNA sequence of a gene. See also:intron
An enzyme that cleaves nucleotides sequentially from free ends of a linear nucleic acid substrate.
tag (EST) A short strand of DNA that is part of cDNA molecule and can act as identifier of a gene. Used in locating and mapping genes. See also:cDNA, sequence tagged site
In genetics, the identification of multiple specific alleles on a person’s DNA to produce a unique identifier for that person. See also:forensics
in situ hybridization (FISH) A Physical mapping approach that uses fluorescein tags to detect hybridization of probes with metaphase chromosomes and with the less-condensed somatic interphase chromatin.
Use of DNA for identification. Some examples of DNA use are to establish paternity in child support cases; establish the presence of a suspect at a crime scene, and identify accident victims.
genomics Study of genes, their resulting proteins, the role played by proteins in the body’s biochemical processes.
See:electrophoresis
Gene gun or particle gun: a popular and widely used direct gene transfer method for delivering
foreign genes into virtually any tissues and cells or even intact seedlings.
• The foreign DNA is coated or precipitated onto the surface of minute gold or tungsten particles (1-3
μm).
• It is bombarded or shot onto the target tissue or cells using the gene gun or microprojectile gun or
shot gun.
• The bombarded cells or tissues are cultured on selection medium to regenerate plants from the
transformed cells.
See:genomic library
Determination of the relative positions of genes on a DNA molecule (chromosome or plasmid) and of the distance, in linkage units or physical units, between them.
All the variations of genes in a species. See also:allele, gene,polymorphism
Experimental procedure aimed at replacing, manipulating, or supplementing nonfunctional or misfunctioning genes with healthy genes. See also:gene, inherit, somatic cell gene therapy, germ line gene therapy
Incorporation of new DNA into an organism’s cells, usually by a vector such as a modified virus. Used in gene therapy. See also:mutation, gene therapy,vector
Altering the genetic material of cells or organisms to enable them to make new substances or perform new functions.
See:recombinant DNA technology
A gene or other identifiable portion of DNA whose inheritance can be followed. See also:chromosome, DNA, gene, inherit Genetic material See:genome Genetic polymorphism Difference in DNA sequence among individuals, groups, or populations (e.g., genes for blue eyes versus brown eyes).
Testing a group of people to identify individuals at high risk of having or passing on a specific genetic disorder.
Analyzing an individual’s genetic material to determine predisposition to a particular health condition or to confirm a diagnosis of genetic disease.
All the genetic material in the chromosomes of a particular organism; its size is generally given as its total number of base pairs.
Research and technology-development effort aimed at mapping and sequencing the genome of human beings and certain model organisms. See also: Human Genome Initiative
A collection of clones made from a set of randomly generated overlapping DNA fragments that represent the entire genome of an organism.
Formerly titled Human Genome Initiative. See also: Human Genome Initiative
Studies performed outside a living organism such as in a laboratory.
Studies carried out in living organisms.
See:genetic marker
A technique for introducing a solution of DNA into a cell using a fine microcapillary pipette or microsyringe under a phase contrast microscope to aid vision. Microsatellite DNA Polymorphism comprising tandem copies of usually, two-, three-, four- or five-nucleotide repeat units, also called a short tandem repeat (STR).
It is an alteration in any of the base of a DNA sequence sometime‘s leading to a defective protein or prematurely terminated non-functional protein. Mutations are spontaneous in nature although rare. Example-Sickle cell haemoglobin has amino acid mutation of valine to glutamine in its beta chain.
Northern blot :- A gel-based laboratory procedure that locates mRNA sequences on a gel that are complementary to a piece of DNA used as a probe.
A virus for which the natural host is a bacterial cell.
Autonomously replicating extra-chromosomal circular DNA molecules, distinct from the normal bacterial genome and non essential for cell survival under nonselective conditions. Some plasmids are capable of integrating into the host genome. Number of artificially constructed plasmids are used as cloning vectors
Enzyme that catalyzes the synthesis of nucleic acids on preexisting nucleic acid templates, assembling RNA from ribonucleotides or DNA from deoxyribonucleotides.
Short preexisting polynucleotide chain(generally from 17-30 nucleotides in length) to which new deoxyribonucleotides can be added by DNA polymerase.
Single-stranded DNA or RNA molecules of specific base sequence, labeled either radioactively or immunologically. Used to detect the complementary base sequence by hybridization.
The nucleotide sequence upstream of a gene that acts as a signal for RNA polymerase binding.
Protein that recognizes specific, short nucleotide sequences and cuts DNA at those sites. Bacteria contain over 400 such enzymes that recognize and cut more than 100 different DNA sequences. See also:restriction enzyme cutting site
Variation between individuals in DNA fragment sizes cut by specific restriction enzymes; polymorphic sequences that result in RFLPs are used as markers on both physical maps and genetic linkage maps. RFLPs are usually caused by mutation at a cutting site. See also:marker, polymorphism
Presence of retroviral vectors, such as some viruses, which use their recombinant DNA to insert their genetic material into the chromosomes of the host’s cells. The virus is then propogated by the host cell.
• A widely used method of determining the order of bases in DNA.
• The classical chain-termination method requires a single-stranded DNA template, a DNA primer, a
DNA polymerase, normal deoxynucleotide triphosphates (dNTPs), and modified nucleotides
(dideoxy NTPs) that terminate DNA strand elongation.
• These ddNTPs will also be radioactively or fluorescently labelled for detection in automated
sequencing machines. The DNA sample is divided into four separate sequencing reactions,
containing all four of the standard deoxynucleotides (dATP, dGTP, dCTP and dTTP) and the DNA
polymerase.
• To each reaction is added only one of the four dideoxynucleotides (ddATP, ddGTP, ddCTP, or
ddTTP) which are the chain-terminating nucleotides, lacking a 3’-OH group required for the
formation of a phosphodiester bond between two nucleotides, thus terminating DNA strand
extension and resulting in DNA fragments of varying length.
See also:sequencing, shotgun sequencing
DNA sequence variations that occur when a single nucleotide (A, T, C, or G) in the genome sequence is
Hereditary disorder caused by a mutant allele of a single gene (e.g., Duchenne muscular dystrophy, retinoblastoma, sickle cell disease). See also:polygenic disorders
Technique Biotechnologist use to create mutation selectively, rather than that which occurs randomly in nature. Using this technique amino acids can be substituted in the expressed proteins making them more stable or functionally better.
Any cell in the body except gametes and their precursors.
Any cell in the body except gametes and their precursors.
Transfer by absorption of DNA fragments separated in electrophoretic gels to membrane filters for detection of specific base sequences by radio-labeled complementary probes.
An experimentally produced organism in which DNA has been artificially introduced and incorporated into the organism’s germ line. See also:cell, DNA, gene, nucleus, germ line
A class of DNA sequences that can move from one chromosomal site to another.
Most common method to introduce rDNA into living cells. In this procedure, bacterial cells take up DNA from the surrounding environment. Many host cell organisms such as E.coli, yeast and mammalian cells do not readily take up foreign DNA and have to be chemically treated to become competent to do so. In 1970, Mandel and Higa found that E.coli cells become markedly competent to take up external DNA when suspended briefly in cold calcium chloride solution. CaCl2 known to increase the efficiency of DNA uptake to produce transformed bacterial cells. The divalent Ca2+ ions supposedly create transient pores on the bacterial cell wall by which the entry of foreign DNA is facilitated into the bacterial cells.
Another method to transfer rDNA into host cells involves mixing the foreign DNA with charged substances like calcium phosphate, cationic liposomes or DEAE dextran and overlaying on recipient animal cells.
DNA molecule, capable of replication in a host organism, into which a gene in inserted to construct a recombinant DNA molecule.
A technique used to identify and locate proteins based on their ability to bind to specific antibodies. See also:DNA, Northern blot, protein, RNA, Southern blotting
Constructed from yeast DNA, it is a vector used to clone large DNA fragments. See also:cloning vector, cosmid
Copyright @ ncerthelp.com A free educational website for CBSE, ICSE and UP board.